- Home
- About
- Partners
- Newcastle University
- University of L'Aquila
- University of Manchester
- Alacris Teranostics GmbH
- University of Pavia
- Polygene
- Consiglio Nazionale delle Ricerche
- INSERM
- Certus Technology
- Charité Universitaet Medizin
- GATC Biotech
- University Medical Center Hamburg Eppendorf
- Evercyte GmbH
- University Hospital of Cologne
- PRIMM Srl
- University of Freiburg
- University of Antwerp
- Finovatis
- Research
- SYBIL at a glance
- Bone
- Growth plate
- Desbuquois dysplasia
- Diastrophic dysplasia
- MCDS
- Osteopetrosis
- Osteoporosis
- Osteogenesis imperfecta
- Prolidase deficiency
- PSACH and MED
- Systems biology
- SOPs
- Alcian Blue staining
- Bone measurements
- BrdU labelling
- Cell counting using ImageJ
- Chondrocyte extraction
- Cre genotyping protocol
- DMMB assay for sulphated proteoglycans
- Densitometry using ImageJ
- Double immunofluorescence
- Electron microscopy of cartilage - sample prep
- Extracting DNA for genotyping
- Grip strength measurement
- Histomorphometry on unon-decalcified bone samples
- Immunocytochemistry
- Immunofluorescence
- Immunohistochemistry
- Quantitative X-ray imaging on bones using Faxitron and ImageJ
- Skeletal preps
- TUNEL assay (Dead End Fluorimetric Kit, Promega)
- Toluidine Blue staining
- Toluidine Blue staining
- Von Kossa Gieson staining
- Wax embedding of cartilage tissue
- Contact Us
- News & Events
- Links
- Portal
Systems biology
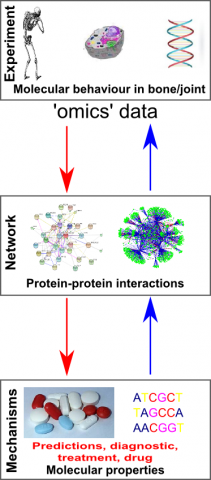 The traditional approaches of biological research are mainly based on experimentation, where observations are used to describe biological phenomena. However, disorders such as skeletal diseases are due to numerous and complex factors, which are difficult to understand through observation alone. At the same time, technological progress has led to a considerable increase in the amount of experimental data that can be collected, opening up the so-called ‘omics’ fields: for example genomics studies the DNA sequences and genes of an organism, proteomics studies all the proteins in a cell type or organism, and metabolomics studies all the small molecules (metabolites) they use. The traditional biology approaches are challenged to cope with this vast amount of data.
The traditional approaches of biological research are mainly based on experimentation, where observations are used to describe biological phenomena. However, disorders such as skeletal diseases are due to numerous and complex factors, which are difficult to understand through observation alone. At the same time, technological progress has led to a considerable increase in the amount of experimental data that can be collected, opening up the so-called ‘omics’ fields: for example genomics studies the DNA sequences and genes of an organism, proteomics studies all the proteins in a cell type or organism, and metabolomics studies all the small molecules (metabolites) they use. The traditional biology approaches are challenged to cope with this vast amount of data.
The system biology approach relies on the combination of experimentation with mathematical modelling and computational tools in order to exploit these new data. The study of interactions is crucial to systems biology: no molecule or process works in isolation in living organisms, but it is their combination that eventually leads to biological function. The link between ‘omics’ datasets and function is established through the construction of networks and mathematical models, which enable us to understand how multiple molecular entities interact which each other to create biological processes.
In the SYBIL project, we are for the first time applying this approach to skeletal diseases. With the help of computational technologies, we are integrating experimental ‘omics’ datasets collected from a variety of disease tissues and cell types into large-scale networks, which will produce a global view of the processes acting in these disorders. These integrated models will enable us to formulate new strategies to combat them, which will eventually be tested by experimentalists towards the development of new drugs.


